Ijraset Journal For Research in Applied Science and Engineering Technology
- Home / Ijraset
- On This Page
- Abstract
- Introduction
- Conclusion
- References
- Copyright
An Overview of Current Research on Computer-Aided Liver Diagnosis Based on Computed Tomography Employing Deep Learning Methods
Authors: Tamilselvan Arjunan
DOI Link: https://doi.org/10.22214/ijraset.2024.58945
Certificate: View Certificate
Abstract
Compared to traditional X-rays, computed tomography (CT) scanning is a non-invasive diagnostic imaging method that yields more precise information about the liver. In contrast to ultrasonography (US) exams, the quality of a CT scan is not heavily operator dependent. Many studies have been conducted with improved outcomes utilizing traditional machine learning methods for computer-aided diagnosis (CAD) of the liver. Recent developments, particularly in the field of deep learning technology, have made it possible to identify, categorize, and segment patterns in medical images. These developments have also been used to other fields outside of medicine. The ability of deep learning to automatically learn feature representations from data, as opposed to feeding in manually created features based on application, is one of its fundamental capabilities. The fundamentals of deep learning are presented in this paper, along with their achievements in liver segmentation and lesion detection, classification utilizing CT imaging modalities, and discussion of their various network topologies. Another fascinating deep learning strategy that is covered is transfer learning. Thus, deep learning and the CAD system have had a significant influence and improved the performance of the healthcare sector.
Introduction
I. INTRODUCTION
Medical imaging techniques like ultrasound, X-ray, computed tomography (CT), magnetic resonance imaging (MRI), positron emission tomography (PET), and mammography are used to create images of parts of the human body for diagnostic and treatment purposes [1,2]. These images are mostly interpreted by radiologists and at the cost of potential fatigue of experts, poor imaging quality and wide variations in pathology, CAD have begun to benefit. Among these imaging techniques, our focus is on CT. In computed tomography, the X-ray beam moves in a circle around the body and that gives many different views of the same organ or structure. The X-ray information is sent to a computer that interprets the X-ray data and displays it in a a two-dimensional (2D) display on a screen. These pictures are kept for future use and diagnosis [3]. A contrasting substance is also used during CT scans to brighten the area being examined. Without the assistance of human professionals, CAD systems automatically diagnose the patient based on image processing and machine learning techniques.
Figure 1 illustrates this. Conventional machine learning techniques, in order to extract informative features from images that explain patterns or regularities in the image and make it hard for non-experts, rely on the domain knowledge of human experts. Deep learning, on the other hand, completes this learning phase automatically, requiring only input data, and it learns the informative representations on its own [4]. The task of feature learning has been transferred from human specialists to computers with improved performance thanks to deep learning. However, in order for deep learning algorithms to be implemented effectively, graphics processing units (GPU) are needed.
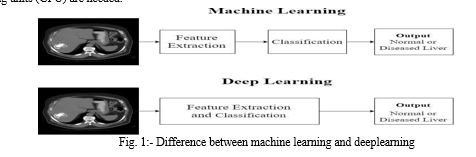
The fundamentals of deep learning are covered in this review, followed by an assessment of how well CAD systems are currently performing in diagnosing liver diseases based on earlier publications. The benefits and issues that need to be resolved for the development of a deep learning-based CAD system as a screening tool for CT examination of liver disease are also covered.
II. DEEP LEARNING
Artificial neural networks, the fundamental algorithm of deep learning, are a branch of machine learning that draw inspiration from the structure and operations of the human brain. Massive amounts of data are required for deep learning algorithms, and augmenting existing data to expand the dataset is known as data augmentation. The first neural network with a single input and output layer was the perceptron [5]. Between the input and output layers of a multi-layer perceptron (MLP), there is an additional layer known as the hidden layer. In neural networks (NN), units inside a layer are not connected to one another, but neighboring layers' units are fully coupled. Feed forward neural networks (FFNN) is another term for MLP. The activation function, bias, and weight of each unit in a layer define it. The non-linear transformation we apply to the input signal is known as the activation function. The following layer of units receives this altered output as input. Deep models are networks that have several hidden layers.
Convolutional neural networks are a subset of deep models that, in contrast to deep Boltzmann machines (DBM), stacked auto encoders (SAE), and deep belief networks (DBN), better exploit the spatial information in images. CNNs are structurally composed of three layers: convolution, pooling, and fully linked. These three deep models—SAE, DBN, and DBM—are used in unsupervised feature representation learning applications, where the input is always in vector form. Other deep network models, such as generative adversarial networks (GAN) and recurrent neural networks (RNN), are briefly covered in [6, 7].
III. CURRENT STATUS OF COMPUTER-AIDED CT LIVER LESION DIAGNOSIS
This review examines and summarizes a number of papers that use deep learning techniques. Better performing CNN-based architectures have been suggested by a number of writers. This paper reports on deep models such as RNN, GAN, and some deep learning based segmentation. It also describes transfer learning approaches employing AlexNet and ResNet.
A. Lesion detection using Perceptual Hash (PH) function
Image hash function also called Image Hashing maps an input image to a value called image hash, which has been used in many applications like image authentication, image copy detection etc. In general the perceptual image hashes are resistant to attacks such as JPEG conversion and geometric transformations. Two basic methods that is durable against these attacks are Discrete Wavelet Transform (DWT) and Singular Value Decomposition (SVD). The perceptual image hashing produces a hashed image using the perceptual hash function.
Ozyurt et al. [8] have developed a method named F- PH-CNN to differentiate between benign and malignant masses from CT liver images. The author has proposed Perceptual Hash (PH) method combined with deep learning and their main objective is to reduce the computational cost using the proposed method. In this work the PH method extracts salient features from the images and also reduces the features by DWT and SVD. The PH function was obtained by applying a 2 level DWT to the input images of fixed size (256*256*3) and singular values were obtained by applying SVD on the divided (2*2) blocks of LL2 and the singular values were normalized. The obtained PH function after applying 2 level DWT was resistant to JPEG compression and applying SVD to LL2 band was resistant to angular attacks [9]. As a result of applying this hash function to 112 CT input images, these images were down sampled to a fixed size of (16*16), (32*32), (64*64), (128*128) by using DWT-SVD perceptual hash which represents the salient features of the images. This DWT-SVD based PH can be said as a pre- processing stage where these fixed size matrices are fed as a vector to the input of the CNN. The CNN has 5 convolutional layers and 2 fully connected layers and the features from the last fully connected layer was fed into ANN, SVM, KNN respectively and the best accuracy was noted. A best accuracy of 98.2% was obtained using the image size of (32*32) with ANN classifier and 9.08s execution time. The author has proposed a method which gave a best accuracy of 98.2% using the image of size only (32*32).
Akif Dogantekin et al. [10] have developed PH-CNN- ELM method to classify benign and malignant masses from Liver CT images. A Perceptual Hash Function (PH) to reduce the size of liver images occupied on hard disk so that the computation time is reduced, a Convolutional Neural Network (CNN) to extract optimal features from the input data which prevents manual feature extraction, an Extreme Learning Machine (ELM) for the classification purpose. The hash function was based on Discrete Wavelet Transform (DWT) to get the LL2 band and Singular Value decomposition (SVD) to get individual values of images where the input images were resized to (256*256*3) and this PH has resulted images with reduced dimensions such as (16*16), (32*32), (64*64), (128*128).
The images were given as input to CNN which has 5 convolution layers and 2 fully connected layers and the features of the last layer were given as inputs to classifiers ELM, SVM, KNN and the best classifier is opted. The data set has been increased to 200 images from available 145 liver CT images using augmentation techniques. A best accuracy of 97.3% was achieved using (32*32) PH image and ELM classifier and the occupied reduction ratio of images in hard disk is 113 times with size of 0,310 MB where without PH the actual storage size is 14,76 MB. The novelty of this work is the PH function which has drastically reduced the execution time of CNN using reduced dimensions of input images and the reduction in memory space.
???????B. Transfer learning approach
Transfer learning is one of the three approaches of deep learning. The other two were classification using a pre- trained model like AlexNet or GoogleNet and then building a network from scratch i.e. training a network ourselves from the start. Transfer learning is an interesting approach where we could use a pre-trained model like AlexNet and modify that network for a different application by making some changes in the network’s architecture [11]. For example, AlexNet is trained for 1000 classes of natural images and we could use that network to classify some other classes other than those 1000 classes by making some modifications in the pre-trained network based on our application.
Weibin Wang et al. [12] have used the residual convolutional neural network (ResNet) for the classification of focal liver lesions (FLL) such as cysts, focal nodular hyperplasia (FNH), hepatocellular carcinoma (HCC), and hemangioma (HEM) using non contrast, arterial and portal venous phase CT images. ResNet eases the training of network where Deep Neural Networks (DNN) poses difficulty in training. The author has shown the significance of fine tuning (Transfer Learning) of networks when only smaller training samples are available. This work has used the available networks AlexNet [13] and ResNet [14] and has fine-tuned the networks for the classification of FLLs. Initially the network has been trained with ImageNet that contains 1 million natural images of 1000 classes and the weights of convolutional layers was saved. The output of the fully connected layer was only changed to 4 class since the original network has 1000 classes. The network was retrained using the medical image data and only the parameters of the fully connected layers were updated. In the pre-processing stage, the ROIs in 388 multi-phase CT images has been extracted by radiologist and the three phases of each image was merged into a three channel image of size (227*227*3) where each channel represents a phase of CT image. These 3 channel images were fed into ResNet for feature extraction which has 49 convolutional layers and 1 fully connected layer. Fine tuning has improved the accuracy of AlexNet from 78.23% to 82.94% and ResNet from 83.67% to 91.22%. This work has managed to achieve better accuracy compared to existing methods for classifying FLLs using fine tuning even when only smaller data is available.
Yu, Y., Wang et al. [15] have proposed a transfer learning approach using AlexNet-CNN for staging liver fibrosis using the biopsy samples of rat liver and the images acquired from SHG microscopy were used after contrast enhancement and adaptive thresholding. The input and output layers of AlexNet was modified according to liver fibrosis assessment and the processed images were resized to 224*224*3 and the output has five stages of fibrosis F0-F4 [16]. This work has also built conventional supervised methods such as artificial neural networks (ANN), multinomial logistic regression (MLR), support vector machines (SVM) and random forests (RF) using 130 morphological and textural features. The results were validated using area under receiver operating characteristic (AUROC) values of up to 0.85–0.95 versus ANN (AUROC of up to 0.87–1.00), MLR (AUROC of up to 0.73–1.00), SVM (AUROC of up to 0.69–0.99) and RF (AUROC of up to 0.94–0.99). The author has defended his work in a way that his transfer learning approach is fully automated and has resulted better accuracy similar to the conventional methods and this transfer learning approach was trained with irrelevant image sources of natural images (AlexNet) to address the requirement of large datasets for deep learning approaches.
???????C. Deep models for liver lesion detection
H. R. Roth et al., [17] have proposed a method for classifying 5 classes of human anatomy such as neck, lungs, liver, pelvis and legs using CT images and ConvNets. The proposed ConvNet has 5 convolutional layers, 2 fully connected layer and a soft max layer which gives the probability for each class and the input images were rescaled to 256*256. Data augmentation was done using random translation, rotations and non-rigid deformations to enrich the data set. The ratio of the training and testing data set was 8:2. This method has resulted in an error rate of 9.6% before augmentation and 5.9% after augmentation and the author has conveyed the importance of data augmentation in error rate reduction.
Amita Das et al., [18] have proposed a new technique called watershed Gaussian based deep learning (WGDL) for automated classification of three types of liver cancer i.e. hemangioma (HEM), hepatocellular carcinoma (HCC) and metastatic carcinoma (MET) using deep neural network (DNN).
The marker controlled watershed segmentation algorithm [19, 20] and morphological operations were used to separate liver region from 225 CT images and Gaussian mixture model (GMM) [21] was used to segment the cancer region from the liver area. The statistical, geometrical and texture features were extracted from the segmented images using gray level co-occurrence matrix (GLCM) method and these features were fed as inputs to the DNN. The training and testing dataset ratio was 7:3 and this method has resulted 99.39% accuracy with DNN. The network parameters can be referred in the paper.
Eugene Vorontsov et al. [22] have evaluated the performance of fully convolutional network (FCN) for detection and segmentation of colorectal liver metastasis (CLM) using CT images. Liver tumour segmentation challenge (LiTS) CT dataset [23] has been used for training and validation and testing dataset of 26 CT images has been taken from Canadian tissue repository network [24]. The model has 2 FCNs and both have U-Net [25] type of architecture. FCN1 results the probability of each pixel of an input image being within the liver and FCN2 outputs the probability of each pixel being a lesion from the FCN1 output. The segmentation result has been compared with the manual segmentation done by the radiologists. The detection sensitivity of automated method for lesion size <10 mm, 10- 20 mm, >20 mm was 10%, 71%, and 85%. Manual correction by the experts after automated segmentation was done and it resulted 57%, 89%, 100% for the same three lesion sizes. The authors had concluded that user correction can generally resolve deficiencies of fully automated segmentation for small metastases and is faster than manual segmentation.
Choi KJ et al. [26] have developed a Liver Fibrosis staging system using deep learning system (DLS) and portal venous phase CT images. Five stages of fibrosis were considered i.e. F0 = no fibrosis, F1 = portal fibrosis, F2 = periportal fibrosis, F3 = septal fibrosis, F4 = cirrhosis. The dataset has 7461 CT examinations for training and 891 CT examinations for testing set. Since the amount of data for F1, F2, and F3 was smaller than F0, F4, the data of stages with lesser data was augmented so that the accuracy is well maintained for minority classes. Liver region of 50 random CT images from the training dataset has been manually segmented by experts and liver segmentation algorithm was developed with five-fold cross validation and this algorithm has resulted in a Dice similarity index of 0.92. Then the entire 7461 CT training set was fed into the segmentation algorithm and then into the convolutional neural network for fibrosis staging. The trained DLS was validated using the test dataset and it has resulted 94%, 95%, 92% accuracy for fibrosis stages F2-F4, F3-F4, F4. Huge collection of images was used in this work with the deep learning technique and has resulted an accurate staging of liver fibrosis.
Avi Ben-Cohen et al. [27] have proposed a liver metastases detection system on global context using FCN and on local context using super-pixel sparse based classification. The local context works on patch level since small metastases are very crucial in early detection of liver cancer. A dataset of 20 patients with 68 lesions for training and 14 patients with 55 lesions for testing was used in which 35% were small lesions since its longest diameter was less than 1.5 cm. The first module was based on FCN-4s architecture using the VGG-16 layer net which produces a lesion probability map. The network was trained with liver image and manually segmented lesion masks by experts. The second module performs local analysis from the FCN network output. For each output from FCN, a super-pixel map was generated where super-pixels [28] reduces the redundancy of densely extracted patches. Each super pixel was represented by a 125 long feature vector and the features from the super-pixels were fed into a sparsity based classifier K-SVD to generate an initial dictionary for lesion detection [29]. In addition the output of FCN was also fed into the classifier for each test case as a fine tuning. Finally the super- pixels were classified as lesion or non-lesion and this work has resulted 94.6% true positive rate and 2.9 false positive rate where false positive reduction will be a promising approach in medical applications.
Yudan Huang et al. [30] have developed a cross- contrast neural network (CCNN) which combines CNN with the modified information based similarity (IBS) [31] method for liver fibrosis classification of five fibrosis stages (F0-F4). This work has used MRI images which are beyond our study but the technique CCNN compares the feature distributions of two images to find whether they come from the same stage of liver fibrosis. The first part of this work has resulted cross probability maps for utilizing the implicit contrast information among the inputs and the second part has measured the similarities i.e. distance between feature vectors between maps using IBS theory. This method has achieved 90% for binary classification, 85% for 4 categories classification on a dataset of only 34 patients, which proves that CCNN can be applied to training relatively small medical dataset for achieving high accuracy.
K. Yasaka, et al. [32] have developed a convolutional neural network (CNN) to classify five classes of liver disease (class 1 - classic hepatocellular carcinomas [HCCs], class 2 - malignant liver tumours other than classic and early HCCs, class 3 - indeterminate masses or mass-like lesions, class 4 - hemangiomas and class 5 - cysts. This work has collected a huge dataset of 1068 image sets from 460 patients for training and that includes all the five classes. Testing dataset has 100 liver masses and all these images includes the three CT phases i.e. non-contrast, arterial, and delayed. Data augmentation [33] was performed on training data and one image has produced 52 image sets and 1068 image sets has produced 55,536 image sets for training.
The test image sets were cropped to the central (500*500) part and all the images was rescaled to (70*70). The CNN was built with six convolutional layers, three max- pooling layers and 3 fully connected layers. The built network has resulted median accuracy of 0.95 and 0.84 for training and test data. The sensitivity of each individual classes for test data were 0.71, 0.33, 0.94, 0.90, and 0.10. The use of GeForce GTX 1080 GPU and 64GB RAM, has allowed them to work on such huge dataset.
Maayan Frid-Adar, el al. [34] have proposed a method for generating synthetic medical images using generative adversarial network (GAN) and liver lesion classification using CNN. The dataset has 53 cysts, 64 metastases and 65 hemangiomas portal-phase 2D CT images and ground-truths were marked by the radiologist. Since the marked lesions were of different sizes, all the marked ROIs were made into a uniform size (64*64) and these ROIs were fed into the CNN input. In order to get a good classification result, a large labelled training dataset is needed. But getting large medical image dataset is complex. So classic data augmentation (CNN-AUG) like translation, rotation, flipping, scaling was done to enlarge the dataset and to reduce the overfitting [35]. In addition two variants of GANs
[36] deep convolutional GAN (DCGAN) [37] and auxiliary classifier GAN (ACGAN) [38] was used to generate synthesized lesion images. The CNN has 3 pairs of convolutional layer followed by max-pooling layer and two fully connected layers ending with a soft-max layer to classify lesions into three classes. The CNN was trained with classic augmentation dataset and yielded 78.6% sensitivity and 88.4% specificity and then using synthetic data augmentation, an improved 85.7% sensitivity and 92.4% specificity was achieved which includes all three classes. This work has utilized NVIDIA GeForce GTX 980 Ti GPU for their training process. The work has proved the efficiency of synthetic augmentation in medical lesion classification systems.
???????D. Deep learning based segmentation approach
M. Ahmad et al. [39] have proposed a Deep Belief Network (DBN-DNN) model [40] that includes unsupervised pre-training and supervised fine tuning for liver segmentation from CT images. In pre-processing stage, the Hounsfield unit
[41] was applied in range [-100,400] to neglect other organs than liver from the CT image and Histogram equalization and Gaussian filtering was done to enhance contrast and to remove noise. The training data were given as (23*23) blocks to the network’s input in which the positive blocks has the region of liver and the negative blocks has the region of background and totally 2,099,712 overlapping blocks was obtained from the training dataset. Usually DBN is composed of stacked Restricted Boltzmann Machines (RBM) and this work has used 2 RBMs for DBN and as a binary classifier sigmoid unit was used and this whole setup was termed as DBN-DNN. The supervised fine tuning was done to reduce the training error using back propagation neural networks. During unsupervised training, only the RBMs were considered and fine tuning was done on all the DBN layers. All the network’s parameters were briefly described in the paper the entire approach has taken 48 hours for the training using CPU. Chen Vase (CV)-based 3D active contour method [42] was applied at DBN_DNN output to refine the liver segmentation by removing any excess noise other than liver region. The author has also validated his method for tumour detection and has achieved good results within the liver region but not at the liver boundary. This work has achieved 94.80% Dice similarity coefficient (DSC) on mixed images and 91.83% DSC on only diseased images.
Ben-Cohen, Avi et al. [43] have explored a FCN for liver segmentation and liver metastases detection using CT images. The dataset includes 20 patients with 68 lesions. This work has used FCN-8s DAG network which uses VGG-16 architecture in which final classifier layer was discarded and fully connected layers were converted to convolutions. The number 8s, 4s in FCN denotes the amount of stride which was briefly described in [44]. Since the dataset was small, for each image four augmentations were created with different scales. Two networks were trained for liver segmentation and lesion detection. The liver segmentation network was trained with 20 CT scans with liver segmentation masks and lesion detection network was trained with 68 lesion segmentation masks and 43 liver segmentation masks. The results were evaluated with two architectural variations: adding two neighbouring slices and linking the Pool2 layer for the final prediction (FCN-4s). A best Dice index of 0.89 and 0.86 sensitivity was obtained using FCN-8s with addition of neighbouring slices for liver segmentation. For lesion detection, the FCN-4s network with addition of neighbouring slices has achieved 0.88 true positive rate (TPR). FCN-4s was more accurate than FCN -8s for lesion detection because of the smaller lesions size. Finally these two best networks for liver segmentation and lesion detection were combined and a fully automated method has achieved 0.86 TPR where the detection results are close to the manual segmentation methods according to the author.
X. Guo, et al. [45] have proposed a transfer learning approach using Mask-RCNN to separate highly clumped steatosis droplets and recover their precise contours with good accuracy from liver histopathological microscopy images. The Mask-RCNN was initially proposed for object instance segmentation [46].
The input colour image was converted to gray scale and then binarized using a normalized threshold to exclude the non-tissue areas and overlapped steatosis regions were identified and removed by rejecting connected foreground regions for solidity over
0.95. Using [47] segmentation masks were created and the dataset has 451 images of size (1024*1024*3) with their corresponding ground-truth masks. Each image has multiple masks where each mask contains one steatosis droplet. So image-mask pairs were given into the network as input. This transfer learning approach has three components: 1) the backbone CNN has three residual neural network (Resnet) modified resnet41, resnet50, resnet65. 2) Region Proposal Network (RPN) for identifying steatosis droplets in input images. 3) ‘ROI-align’ analyses the ROI and interpolates the feature map from backbone CNN. As a result modified resnet50 has achieved a best precision of 75.87% and a Jaccard index of 76.97%.
IV. DISCUSSION
As discussed all through this review, deep learning has the advantage of learning the feature representation from the input data by itself without the need for hand crafted features using conventional methods. This survey has discussed various deep learning architectures like CNN, RNN, CCNN, GAN, and DBN for the task of liver lesion detection using CT images. These architectures have used various numbers of different layers like convolution, pooling, fully connected layers based on the application’s objective. Usually deep learning is said to process huge amount of data but getting huge collection of medical image data is complex. So data augmentation has become so popular in building deep learning networks for medical image diagnosis. These data augmentation techniques help the deep learning networks to avoid overfitting of data and in improving data strength. So data augmentation techniques make way for deep learning implementation for medical diagnosis using small amount of data. A comparison is presented below in Table 1. Deep learning algorithms with huge data is very difficult for CPUs to handle and most of the works have opted for GPUs like NVIDIA’s Tesla, Titan, GeForce products. These graphics processors greatly reduces the training time of the networks and deep learning on GPUs is typically 10–30 times faster than on CPUs. The memory size of the GPUs also poses some limitations on the batch size of the training data [30]. Some of the GPUs that has been mentioned in the above survey is given in table 2 and as evident more no of classes has been classified in most of the works that has used GPU acceleration.
Table 1:- Comparison between different augmentation techniques listed in this review
|
Approach |
Augmentation Technique |
No of images |
Results after Augmentation |
Reference |
|
Perceptual hashing-CNN |
NA |
F From 145 to 200 images |
97.3% |
[10] |
|
ConvNet |
Random translation, rotations and non-rigid deformations |
4298 images |
Error rate of 5.9% from 9.6% |
[17] |
|
CNN |
Cropping, rotation, Gaussian noise added |
From 1068 to 55,536 images |
95% accuracy |
[32] |
|
GAN |
Translation, rotation, flipping, scaling and synthetic images |
NA |
85.7% sensitivity and 92.4% specificity |
[34] |
|
FCN |
Scaling (Four scales from 0.8 to 0.12) |
NA |
0.86 TPR |
[43] |
* NA – not available
Table 2:- Different GPUs mentioned in the literature
|
Approach |
GPU |
No of classes |
Reference |
|
ConvNet |
NVIDIA Titan Z |
5 class |
[17] |
|
FCN |
NVIDIA GeForce GTX 1080 |
1 class |
[27] |
|
CNN |
NVIDIA GeForce GTX 1080 |
5 class |
[32] |
|
GAN |
NVIDIA GeForce GTX 980Ti |
3 class |
[34] |
|
CNN |
NVIDIA Tesla K80 |
1 class |
[45] |
|
CCNN |
Nvidia Tesla V100 GPU |
5 class |
[30] |
Transfer learning makes the training process little easier where pre-trained models like AlexNet are just modified in terms of layers based on our application and reused to perform the new task. In case where transfer learning fails to provide the expected result we should build our network from scratch. Most of the works in this review has been implemented using Python and Matlab. Unlike conventional machine learning methods where features are extracted manually based on the application and then fed into the classifier, deep learning algorithms learns features on its own in each layer which could be interpreted or visualized for inference. These layers repeatedly learn features upon iterations until the network converges to the best training accuracy. A similar survey [48] has reviewed all the proposed deep learning techniques for liver lesion detection using ultrasound images. An overview of the above mentioned techniques and proposed methodologies have been portrayed above in figure 2. First division lists the different pre-trained models used for transfer learning approach. Second division lists the different deep learning architectures proposed for our application. Third division lists the utilized Matlab and Python based libraries and the last division lists the publicly available Liver datasets.
 ???????
???????
Conclusion
It is clear from the publications examined in this survey that deep learning is ingrained in CAD systems for the identification of liver lesions. Deep learning approaches have covered every facet of medical image processing, not just the liver region [7] [49]. When it comes to diagnosing liver lesions, conventional approaches have reached about 99% accuracy, but deep learning techniques have not yet reached a high standard. Deep learning approaches can be utilized to treat a variety of scattered liver illnesses that have little discrimination capabilities and are visually identical [50]. For liver segmentation, a large number of these lesion detection algorithms were still user-dependent, and there were still fewer fully automatic deep learning-based segmentation techniques. GPUs are essential to applying deep learning techniques quickly, but their cost is a concern. Despite the complexity of obtaining medical pictures, deep learning approaches are producing better outcomes with less data.
References
[1] Brody H. 2013. Medical imaging. Nature Vol 502 S81, 2013, Issue No 7473. [2] Bernard E. Van Beers, Jean-Luc Daire, Philippe Garteiser, “New imaging techniques for liver diseases,” Journal of Hepatology, vol. 62, pp. 690-700, 2015. [3] John Hopkins. 2019.Medicine, https://www.hopkinsmedicine.org/health/treatment- tests-and therapies/computed-tomography-ct-or-cat- scan-of-the-liver-and- biliary-tract [4] Schmidhuber J. 2015. Deep learning in neural networks: an overview. Neural Netw. 61:85–117 [5] Bengio Y. 2009. Learning Deep Architectures for AI: Foundations and Trends in Machine Learning. Boston: Now. 127 pp. [6] https://medium.com/tensorflow/mit-deep-learning- basics- introduction-and-overview-with-tensorflow- 5bcd26baf0 [7] Shen, D., Wu, G., & Suk, H. (2017). Deep Learning in Medical Image Analysis. Annual review of biomedical engineering, 19, 221-248. [8] Ozyurt, F., Tuncer, T., Avci, E. et al. Arab J Sci Eng (2019) 44: 3173. [9] Tang, Z.; Zhang, X.; Dai, X.; Yang, J.; Wu, T.: Robust image hash function using local colour features. AEU Int. J. Electron. Commun.67 (8), 717–722 (2013). [10] Akif Do?antekin, Fatih Ozyurt, Engin Avc?, Mustafa Koç, A novel approach for liver image classification: PH-C-ELM, Measurement, Volume 137, 2019, Pages332-338, ISSN 0263-224. [11] H. Shin et al., \"Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning,\" in IEEE Transactions on Medical Imaging, vol. 35, no. 5, pp. 1285-1298, May 2016. Doi: 10.1109/TMI.2016.2528162 [12] Weibin Wang et al., 2018. Classification of Focal Liver Lesions Using Deep Learning with Fine-Tuning. In Proceedings of the 2018 International Conference on Digital Medicine and Image Processing (DMIP \'18).ACM, New York, NY, USA, 56-60 [13] Krizhevsky, A., Sutskever, I., Hinton, G. E. Imagenet classification with deep convolutional neural networks, Advances in Neural Information Processing Systems. pp. 1097-1105, 2012. [14] He, K. et al. Deep residual learning for image recognition. Proceedings of 2016 the IEEE conference on computer vision and pattern recognition, pp.770- 778, 2016. [15] Yu, Y., Wang, J., Ng, C.W., Ma, Y., Mo, S., Fong, E.L., Xing, J., Song, Z., Xie, Y., Si, K., Wee, A., Welsch, R.E., So, P.T., & Yu, H. (2018). Deep learning enables automated scoring of liver fibrosis stages. Scientific Reports. [16] Desautels, T. et al. Using Transfer Learning for Improved Mortality Prediction in a Data-Scarce Hospital Setting. Biomedical informatics insights 9, 1178222617712994, https://doi.org/10.1177/1178222617712994 (2017). [17] H. R. Roth et al., ‘‘Anatomy-specific classification of medical images using deep convolutional nets,’’ in Proc. IEEE 12th Int. Symp. Biomed. Imag. (ISBI), Apr. 2015, pp. 101–104. [18] Amita Das et al., ‘Deep learning based liver cancer detection using watershed transform and Gaussian mixture model techniques’, Cognitive Systems Research, Volume 54, 2019, Pages 165-175. [19] Chattaraj, A., Das, A., & Bhattacharya, M. (2017). Mammographic image segmentation by marker controlled watershed algorithm. IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 1000–1003. [20] Masoumi, H., Behrad, A., Pourmina, M. A., & Roosta, (2012). Automatic liver segmentation in MRI images using an iterative watershed algorithm and artificial neural network. Biomedical Signal Processing and Control, 7, 429–437. [21] Kermani, S., Samadzadehaghdam, N., & EtehadTavakol, M. (2015). Automatic colour segmentation of breast infrared images using a Gaussian Mixture Model. Optik, 126, 3288–3294. [22] Eugene Vorontsov et al. Radiology: Artificial Intelligence Volume 1: Number 2—2019 [23] LiTS: Liver Tumour Segmentation challenge. https://competitions.codalab. Org/competitions/17094. Accessed April 28, 2018. [24] Canadian Tissue RepositoryNetwork. http://www.ctrnet.ca/. Accessed January 24, 2019. [25] Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, eds. Medical image computing and computer-assisted intervention – MICCAI 2015. MICCAI 2015. Lecture notes in computer science, vol 9351. Cham, Switzerland: Springer, 2015. [26] Choi KJ, Jang JK, Lee SS, et al. “Development and validation of a deep learning system for staging liver fibrosis by using contrast agent-enhanced CT images in the liver”, Radiology, vol.289, pp. 688–697, 2018. [27] Avi Ben-Cohen et al., “Fully convolutional network and sparsity-based dictionary learning for liver lesion detection in CT examinations”, Neurocomputing, Volume 275, pp. 1585-1594, 2018. [28] R. Achanta, A. Shaji, K. Smith, A. Lucchi, P. Fua, S. Süsstrunk, SLIC superpix- els compared to state-of- the-art superpixel methods, IEEE Trans. Pattern Anal. Mach. Intell. 34 (11) (2012) 2274–2282, doi: 10.1109/TPAMI.2012.120. [29] Z. Jiang, Z. Lin, L.S. Davis, Learning a discriminative dictionary for sparse coding via label consistent K- SVD, in: Proceedings of 2011 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, 2011, pp. 1697–1704, doi: 10.1109/CVPR.2011.5995354 . [30] Yudan Huang, Ying Chen, Haochuan Zhu, Weifeng Li, Yun Ge, Xiaolin Huang, Jian He, A liver fibrosis staging method using cross-contrast network, Expert Systems with Applications, Volume 130, 2019, Pages 124-131, ISSN 0957-4174. [31] Yang, A et al., (2003). Linguistic analysis of the human heartbeat using frequency and rank order statistics. Physical Review Letters, 90 (10), 108103. [32] K. Yasaka, H. Akai, O. Abe, and S. Kiryu, “Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast- enhanced CT: a preliminary study,” Radiology, vol. 286, no. 3, pp. 887–896, 2017. [33] Krizhevsky A, Sutskever I, Hinton G. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems 25 (NIPS 2012). http://papers.nips.cc/paper/4824- imagenetclassification-with-deep- convolutional- neuralnetworks. Published 2012. Accessed January 31, 2017. [34] Maayan Frid-Adar, el al., “GAN-based synthetic medical image augmentation for increased CNN performance in liver lesion classification”, Neurocomputing, Volume 321, 2018, Pages321-331. [35] Krizhevsky , I. Sutskever , G.E. Hinton ,Imagenet classification with deep convolutional neural networks, in: Proceedings of the Advances in Neural Infor- mation Processing Systems, 2012, pp. 1097–1105 . [36] Goodfellow , J. Pouget-Abadie , M. Mirza , B. Xu , D. Warde- Farley , S. Ozair , A. Courville , Y. Bengio , Generative adversarial nets, in: Proceedings of the Ad- vances in Neural Information Processing Systems, 2014, pp. 2672–2680 . [37] Radford, L. Metz, S. Chintala, Unsupervised representation learning with deep convolutional generative adversarial networks, arXiv:1511.06434 (2015). [38] Odena, C. Olah, J. Shlens, Conditional image synthesis with auxiliary classi- fier gans, arXiv:1610.09585 (2016). [39] M. Ahmad et al., \"Deep Belief Network Modeling for Automatic Liver Segmentation,\" in IEEE Access, vol. 7, pp. 20585- 20595, 2019. doi:10.1109/ACCESS.2019.2896961 [40] G. E. Hinton, S. Osindero, and Y.-W. Teh, ``A fast learning algorithm for deep belief nets,\'\' Neural Comput., vol. 18, no. 7, pp. 1527_1554, 2006. [41] L. R. Dice, ``Measures of the amount of ecologic association between species,\'\' Ecology, vol. 26, no. 3, pp. 297_302, 1945. [42] T. F. Chan and L. A. Vese, ``Active contours without edges,\'\' IEEE Trans. Image Process., vol. 10, no. 2, pp. 266_277, Feb. 2001. [43] Ben-Cohen, Avi et al. “Fully Convolutional Network for Liver Segmentation and Lesions Detection.” LABELS/DLMIA@MICCAI (2016). [44] Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3431– 3440 (2015). [45] X. Guo, F. Wang, G. Teodoro, A. B. Farris and J. Kong, \"Liver Steatosis Segmentation with Deep Learning Methods,\" 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 2019, pp. 24-27. [46] He, K.M. and Gkioxari, G. and Doll´ar, P. and Girshick, R., “Mask R-CNN,” IEEE International Conference on Computer Vision, pp.2980–2988, 2017. [47] Guo, X.Y. and Yu, H.Y. and Rossetti, B. and Teodoro, G. and Brat, D.J. and Kong, J., “Clumped Nuclei Segmentation with Adjacent Point Match and Local Shape based Intensity Analysis for Overlapped Nuclei in Fluorescence In- Situ Hybridization Images,” IEEE International Conference on Engineering in Medicine and Biology, pp.3410–3413, 2018. [48] Nishida, N., Yamakawa, M., Shiina, T. et al. Hepatol Int (2019) 13: 416. https://doi.org/10.1007/s12072-019-09937-4. [49] Geert Litjens, Thijs Kooi, Babak Ehteshami Bejnordi, Arnaud Arindra Adiyoso Setio, Francesco Ciompi, Mohsen Ghafoorian, Jeroen A.W.M. van der Laak, Bram van Ginneken, Clara I. Sánchez, A survey on deep learning in medical image analysis, Medical Image Analysis, Volume 42, 2017, Pages 60-88, ISSN1361-8415. [50] Raghesh Krishnan K, SudhakarRadhakrishnan, “Focal and diffused liver disease classification from ultrasound images based on isocontour segmentation,” IET Image Process., Vol. 9, Iss. 4, pp. 261– 270, 2015.
Copyright
Copyright © 2024 Tamilselvan Arjunan. This is an open access article distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.

Download Paper
Paper Id : IJRASET58945
Publish Date : 2024-03-12
ISSN : 2321-9653
Publisher Name : IJRASET
DOI Link : Click Here
 Submit Paper Online
Submit Paper Online

